EPS format
EPS format
To save these results right click and hit Save As...
propka3.1 2012-08-22
-------------------------------------------------------------------------------------------------------
-- --
-- PROPKA: A PROTEIN PKA PREDICTOR --
-- --
-- VERSION 1.0, 04/25/2004, IOWA CITY --
-- BY HUI LI --
-- --
-- VERSION 2.0, 11/05/2007, IOWA CITY/COPENHAGEN --
-- BY DELPHINE C. BAS AND DAVID M. ROGERS --
-- --
-- VERSION 3.0, 01/06/2011, COPENHAGEN --
-- BY MATS H.M. OLSSON AND CHRESTEN R. SONDERGARD --
-- --
-- VERSION 3.1, 07/01/2011, COPENHAGEN --
-- BY CHRESTEN R. SONDERGARD AND MATS H.M. OLSSON --
-------------------------------------------------------------------------------------------------------
-------------------------------------------------------------------------------------------------------
References:
Very Fast Empirical Prediction and Rationalization of Protein pKa Values
Hui Li, Andrew D. Robertson and Jan H. Jensen
PROTEINS: Structure, Function, and Bioinformatics 61:704-721 (2005)
Very Fast Prediction and Rationalization of pKa Values for Protein-Ligand Complexes
Delphine C. Bas, David M. Rogers and Jan H. Jensen
PROTEINS: Structure, Function, and Bioinformatics 73:765-783 (2008)
PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa predictions
Mats H.M. Olsson, Chresten R. Sondergard, Michal Rostkowski, and Jan H. Jensen
Journal of Chemical Theory and Computation, 7(2):525-537 (2011)
Improved Treatment of Ligands and Coupling Effects in Empirical Calculation
and Rationalization of pKa Values
Chresten R. Sondergaard, Mats H.M. Olsson, Michal Rostkowski, and Jan H. Jensen
Journal of Chemical Theory and Computation, (2011)
-------------------------------------------------------------------------------------------------------
--------- ----- ------ --------------------- -------------- -------------- --------------
DESOLVATION EFFECTS SIDECHAIN BACKBONE COULOMBIC
RESIDUE pKa BURIED REGULAR RE HYDROGEN BOND HYDROGEN BOND INTERACTION
--------- ----- ------ --------- --------- -------------- -------------- --------------
ASP 19 A 3.87 0 % 0.23 208 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X -0.07 N+ 4 A
ASP 19 A 0.00 XXX 0 X 0.00 XXX 0 X -0.08 LYS 18 A
ASP 19 A 0.00 XXX 0 X 0.00 XXX 0 X -0.01 HIS 17 A
ASP 32 A 3.58 25 % 1.22 350 0.12 0 -0.80 THR 108 A -0.02 ILE 33 A -0.29 LYS 111 A
ASP 32 A 0.00 XXX 0 X -0.71 LYS 111 A -0.02 LYS 113 A
ASP 32 A 0.00 XXX 0 X 0.00 XXX 0 X 0.13 ASP 34 A
ASP 32 A 0.00 XXX 0 X 0.00 XXX 0 X -0.01 HIS 107 A
ASP 32 A 0.00 XXX 0 X 0.00 XXX 0 X 0.15 ASP 110 A
ASP 34 A 3.33 0 % 0.38 247 0.00 0 0.00 XXX 0 X -0.48 HIS 36 A -0.08 LYS 111 A
ASP 34 A 0.00 XXX 0 X -0.33 THR 37 A -0.17 HIS 36 A
ASP 34 A 0.00 XXX 0 X 0.00 XXX 0 X 0.21 ASP 110 A
ASP 41 A 2.04 0 % 0.56 232 0.00 0 -0.85 SER 43 A -0.81 SER 43 A -0.10 LYS 39 A
ASP 41 A -0.17 LYS 257 A -0.00 LEU 44 A -0.04 LYS 45 A
ASP 41 A 0.00 XXX 0 X 0.00 XXX 0 X -0.34 LYS 257 A
ASP 52 A 3.50 0 % 0.29 226 0.00 0 -0.09 GLN 53 A -0.38 ASP 52 A -0.15 ARG 182 A
ASP 52 A 0.00 XXX 0 X 0.00 XXX 0 X 0.04 ASP 180 A
ASP 71 A 1.68 1 % 0.45 285 0.00 0 -0.66 SER 73 A -0.72 SER 73 A -0.04 ARG 89 A
ASP 71 A -0.77 LYS 76 A 0.00 XXX 0 X -0.38 LYS 76 A
ASP 72 A 4.21 33 % 1.14 374 0.18 0 -0.66 TRP 123 A -0.25 ASP 72 A -0.14 ARG 89 A
ASP 72 A 0.00 XXX 0 X 0.00 XXX 0 X 0.13 ASP 71 A
ASP 72 A 0.00 XXX 0 X 0.00 XXX 0 X 0.02 ASP 130 A
ASP 75 A 3.33 12 % 0.75 316 0.00 0 -0.77 ARG 89 A 0.00 XXX 0 X -0.02 LYS 80 A
ASP 75 A 0.00 XXX 0 X 0.00 XXX 0 X -0.43 ARG 89 A
ASP 85 A 3.82 0 % 0.19 258 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X -0.14 LYS 45 A
ASP 85 A 0.00 XXX 0 X 0.00 XXX 0 X -0.07 LYS 127 A
ASP 85 A 0.00 XXX 0 X 0.00 XXX 0 X 0.05 ASP 139 A
ASP 101 A 3.14 11 % 0.54 311 0.00 0 -0.84 ARG 227 A 0.00 XXX 0 X -0.37 ARG 227 A
ASP 110 A 2.44 0 % 0.60 254 0.00 0 -0.85 THR 35 A -0.84 THR 35 A -0.03 LYS 111 A
ASP 110 A 0.00 XXX 0 X 0.00 XXX 0 X -0.25 HIS 36 A
ASP 130 A 3.59 0 % 0.41 217 0.00 0 0.00 XXX 0 X -0.02 GLY 132 A -0.16 LYS 133 A
ASP 130 A 0.00 XXX 0 X -0.44 LYS 133 A 0.00 XXX 0 X
ASP 139 A 2.36 13 % 0.74 318 0.00 0 -0.47 LYS 127 A -0.43 ASP 139 A -0.04 ARG 27 A
ASP 139 A -0.85 TYR 128 A 0.00 XXX 0 X -0.39 LYS 127 A
ASP 162 A 3.45 0 % 0.39 268 0.00 0 -0.53 GLN 158 A 0.00 XXX 0 X -0.21 LYS 225 A
ASP 165 A 3.84 0 % 0.21 263 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X -0.06 LYS 168 A
ASP 165 A 0.00 XXX 0 X 0.00 XXX 0 X -0.11 LYS 225 A
ASP 165 A 0.00 XXX 0 X 0.00 XXX 0 X -0.05 LYS 228 A
ASP 165 A 0.00 XXX 0 X 0.00 XXX 0 X 0.05 ASP 162 A
ASP 175 A 3.92 0 % 0.14 191 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X -0.02 ARG 58 A
ASP 180 A 2.35 14 % 0.89 320 0.06 0 -1.29 ARG 182 A -0.68 ARG 182 A -0.04 LYS 159 A
ASP 180 A 0.00 XXX 0 X 0.00 XXX 0 X -0.38 ARG 182 A
ASP 190 A 3.98 3 % 0.43 291 0.01 0 -0.01 SER 188 A 0.00 XXX 0 X -0.27 LYS 213 A
ASP 190 A 0.00 XXX 0 X 0.00 XXX 0 X 0.04 GLU 214 A
ASP 243 A 2.52 38 % 1.47 387 0.10 0 -0.74 SER 99 A -0.52 SER 99 A -0.07 ARG 227 A
ASP 243 A -0.81 GLN 103 A 0.00 XXX 0 X 0.00 XXX 0 X
ASP 243 A -0.71 TRP 245 A 0.00 XXX 0 X 0.00 XXX 0 X
GLU 14 A 3.84 0 % 0.44 260 0.00 0 0.00 XXX 0 X -0.62 GLU 14 A -0.10 LYS 9 A
GLU 14 A 0.00 XXX 0 X 0.00 XXX 0 X -0.04 LYS 18 A
GLU 14 A 0.00 XXX 0 X 0.00 XXX 0 X -0.05 HIS 10 A
GLU 14 A 0.00 XXX 0 X 0.00 XXX 0 X -0.28 HIS 15 A
GLU 26 A 4.50 0 % 0.22 184 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X -0.26 LYS 252 A
GLU 26 A 0.00 XXX 0 X 0.00 XXX 0 X -0.04 ARG 254 A
GLU 26 A 0.00 XXX 0 X 0.00 XXX 0 X 0.08 GLU 205 A
GLU 69 A 4.88 33 % 1.32 375 0.16 0 -0.66 ARG 58 A 0.00 XXX 0 X -0.44 ARG 58 A
GLU 106 A 5.12 100 % 4.23 652 1.01 0 -0.85 THR 199 A -0.84 ARG 246 A -3.08 ZN ZN A
GLU 106 A 0.00 XXX 0 X 0.00 XXX 0 X -0.29 ARG 246 A
GLU 106 A 0.00 XXX 0 X 0.00 XXX 0 X -0.52 HIS 107 A
GLU 106 A 0.00 XXX 0 X 0.00 XXX 0 X 0.95 GLU 117 A
GLU 117 A 3.18 100 % 4.09 659 0.60 0 -1.60 HIS 107 A -0.71 GLU 106 A -1.15 ZN ZN A
GLU 117 A 0.00 XXX 0 X -0.79 HIS 107 A -1.76 HIS 107 A
GLU 187 A 4.83 0 % 0.10 129 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.23 GLU 214 A
GLU 205 A 3.96 28 % 1.22 359 0.13 0 -0.75 GLN 28 A -0.75 ARG 27 A -0.19 ARG 27 A
GLU 205 A 0.00 XXX 0 X 0.00 XXX 0 X -0.02 ARG 246 A
GLU 205 A 0.00 XXX 0 X 0.00 XXX 0 X -0.06 LYS 252 A
GLU 205 A 0.00 XXX 0 X 0.00 XXX 0 X -0.11 ARG 254 A
GLU 214 A 3.75 2 % 0.60 288 0.02 0 0.00 XXX 0 X -0.51 GLU 187 A -0.04 LYS 149 A
GLU 214 A 0.00 XXX 0 X -0.71 SER 188 A -0.08 LYS 154 A
GLU 214 A 0.00 XXX 0 X 0.00 XXX 0 X -0.03 LYS 213 A
GLU 221 A 3.89 0 % 0.23 195 0.00 0 -0.20 SER 219 A -0.60 GLU 221 A -0.03 LYS 225 A
GLU 234 A 3.64 0 % 0.21 171 0.00 0 -0.68 LYS 172 A 0.00 XXX 0 X -0.38 LYS 172 A
GLU 236 A 3.87 0 % 0.25 203 0.00 0 0.00 XXX 0 X -0.83 GLY 233 A -0.05 LYS 170 A
GLU 238 A 3.22 0 % 0.33 234 0.00 0 -0.85 ASN 230 A 0.00 XXX 0 X -0.34 LYS 168 A
GLU 238 A -0.42 LYS 168 A 0.00 XXX 0 X 0.00 XXX 0 X
GLU 239 A 5.15 20 % 0.60 336 0.02 0 0.00 XXX 0 X 0.00 XXX 0 X -0.01 LYS 170 A
GLU 239 A 0.00 XXX 0 X 0.00 XXX 0 X 0.03 GLU 236 A
HIS 10 A 6.39 0 % -0.14 115 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X -0.01 LYS 18 A
HIS 10 A 0.00 XXX 0 X 0.00 XXX 0 X 0.05 GLU 14 A
HIS 15 A 6.26 0 % -0.42 211 0.00 0 0.00 XXX 0 X 0.41 LYS 9 A -0.03 LYS 9 A
HIS 15 A 0.00 XXX 0 X 0.00 XXX 0 X -0.38 LYS 18 A
HIS 15 A 0.00 XXX 0 X 0.00 XXX 0 X -0.10 HIS 10 A
HIS 15 A 0.00 XXX 0 X 0.00 XXX 0 X 0.28 GLU 14 A
HIS 17 A 5.54 0 % -0.40 238 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X -0.15 LYS 18 A
HIS 17 A 0.00 XXX 0 X 0.00 XXX 0 X -0.35 LYS 24 A
HIS 17 A 0.00 XXX 0 X 0.00 XXX 0 X -0.07 HIS 15 A
HIS 17 A 0.00 XXX 0 X 0.00 XXX 0 X 0.01 ASP 19 A
HIS 36 A 6.74 0 % -0.18 157 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.17 ASP 34 A
HIS 36 A 0.00 XXX 0 X 0.00 XXX 0 X 0.25 ASP 110 A
HIS 64 A 3.47 87 % -2.29 526 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X -0.45 ZN ZN A
HIS 64 A 0.00 XXX 0 X 0.00 XXX 0 X -0.29 LYS 170 A
HIS 94 A -2.19 100 % -2.94 647 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X -4.07 ZN ZN A
HIS 94 A 0.00 XXX 0 X 0.00 XXX 0 X -0.38 HIS 64 A
HIS 94 A 0.00 XXX 0 X 0.00 XXX 0 X -1.30 HIS 96 A
HIS 96 A -0.62 100 % -3.56 657 0.00 0 0.00 XXX 0 X 0.84 ASN 244 A -4.07 ZN ZN A
HIS 96 A 0.00 XXX 0 X 0.00 XXX 0 X -0.26 HIS 64 A
HIS 96 A 0.00 XXX 0 X 0.00 XXX 0 X -0.08 HIS 107 A
HIS 107 A 6.50 100 % -3.52 650 0.00 0 1.60 GLU 117 A 0.00 XXX 0 X -0.32 ZN ZN A
HIS 107 A 0.00 XXX 0 X 0.00 XXX 0 X -0.05 ARG 254 A
HIS 107 A 0.00 XXX 0 X 0.00 XXX 0 X 0.01 ASP 32 A
HIS 107 A 0.00 XXX 0 X 0.00 XXX 0 X 0.52 GLU 106 A
HIS 107 A 0.00 XXX 0 X 0.00 XXX 0 X 1.76 GLU 117 A
HIS 119 A -4.89 100 % -3.80 698 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X -4.07 ZN ZN A
HIS 119 A 0.00 XXX 0 X 0.00 XXX 0 X -1.24 HIS 94 A
HIS 119 A 0.00 XXX 0 X 0.00 XXX 0 X -1.59 HIS 96 A
HIS 119 A 0.00 XXX 0 X 0.00 XXX 0 X -0.69 HIS 107 A
HIS 122 A 3.58 100 % -3.50 627 0.00 0 0.00 XXX 0 X 0.14 LEU 120 A 0.00 XXX 0 X
HIS 122 A 0.00 XXX 0 X 0.45 ALA 142 A 0.00 XXX 0 X
CYS 206 A 11.58 77 % 2.92 498 0.00 0 0.00 XXX 0 X -0.17 CYS 206 A -0.19 ARG 27 A
CYS 206 A 0.00 XXX 0 X 0.00 XXX 0 X 0.02 GLU 205 A
TYR 7 A 13.35 100 % 3.53 580 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X -1.19 ZN ZN A
TYR 7 A 0.00 XXX 0 X 0.00 XXX 0 X -0.09 ARG 246 A
TYR 7 A 0.00 XXX 0 X 0.00 XXX 0 X 1.10 GLU 106 A
TYR 40 A 10.36 0 % 0.28 180 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.07 ASP 41 A
TYR 40 A 0.00 XXX 0 X 0.00 XXX 0 X 0.01 ASP 190 A
TYR 51 A 13.81 100 % 3.81 597 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.00 XXX 0 X
TYR 88 A 12.05 58 % 1.98 444 0.00 0 -0.37 ASN 124 A 0.00 XXX 0 X -0.00 ARG 27 A
TYR 88 A 0.00 XXX 0 X 0.00 XXX 0 X 0.12 ASP 85 A
TYR 88 A 0.00 XXX 0 X 0.00 XXX 0 X 0.35 ASP 139 A
TYR 88 A 0.00 XXX 0 X 0.00 XXX 0 X 0.00 CYS 206 A
TYR 88 A 0.00 XXX 0 X 0.00 XXX 0 X -0.17 LYS 127 A
TYR 88 A 0.00 XXX 0 X 0.00 XXX 0 X 0.14 TYR 128 A
TYR 114 A 11.04 32 % 1.02 372 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.02 ASP 110 A
TYR 128 A 10.78 0 % 0.63 263 0.00 0 -0.17 GLN 137 A 0.00 XXX 0 X -0.38 LYS 127 A
TYR 128 A -0.54 LYS 127 A 0.00 XXX 0 X 0.39 ASP 139 A
TYR 128 A 0.85 ASP 139 A 0.00 XXX 0 X 0.00 XXX 0 X
TYR 191 A 12.05 38 % 1.97 387 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.06 ASP 41 A
TYR 191 A 0.00 XXX 0 X 0.00 XXX 0 X 0.02 ASP 190 A
TYR 191 A 0.00 XXX 0 X 0.00 XXX 0 X 0.01 TYR 40 A
TYR 194 A 13.92 100 % 4.79 623 0.00 0 -0.85 SER 29 A 0.00 XXX 0 X -0.04 ARG 27 A
TYR 194 A 0.00 XXX 0 X 0.00 XXX 0 X 0.18 GLU 106 A
TYR 194 A 0.00 XXX 0 X 0.00 XXX 0 X 0.40 GLU 117 A
TYR 194 A 0.00 XXX 0 X 0.00 XXX 0 X -0.04 ARG 246 A
TYR 194 A 0.00 XXX 0 X 0.00 XXX 0 X -0.52 ARG 254 A
LYS 9 A 10.55 0 % -0.06 85 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.10 GLU 14 A
LYS 18 A 10.26 0 % -0.36 181 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.04 GLU 14 A
LYS 18 A 0.00 XXX 0 X 0.00 XXX 0 X 0.08 ASP 19 A
LYS 24 A 10.24 0 % -0.23 216 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X -0.00 ARG 246 A
LYS 24 A 0.00 XXX 0 X 0.00 XXX 0 X -0.04 LYS 252 A
LYS 39 A 10.26 0 % -0.13 151 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.10 ASP 41 A
LYS 39 A 0.00 XXX 0 X 0.00 XXX 0 X -0.21 LYS 257 A
LYS 45 A 10.51 0 % -0.18 197 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.04 ASP 41 A
LYS 45 A 0.00 XXX 0 X 0.00 XXX 0 X 0.14 ASP 85 A
LYS 76 A 11.27 0 % -0.38 230 0.00 0 0.77 ASP 71 A 0.00 XXX 0 X -0.00 ARG 89 A
LYS 76 A 0.00 XXX 0 X 0.00 XXX 0 X 0.38 ASP 71 A
LYS 80 A 10.34 0 % -0.18 240 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.02 ASP 75 A
LYS 111 A 10.74 0 % -0.16 215 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.29 ASP 32 A
LYS 111 A 0.00 XXX 0 X 0.00 XXX 0 X 0.08 ASP 34 A
LYS 111 A 0.00 XXX 0 X 0.00 XXX 0 X 0.03 ASP 110 A
LYS 112 A 9.97 10 % -0.50 308 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X -0.03 LYS 149 A
LYS 113 A 10.22 5 % -0.29 296 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.02 ASP 32 A
LYS 113 A 0.00 XXX 0 X 0.00 XXX 0 X -0.02 LYS 111 A
LYS 127 A 12.13 0 % -0.39 256 0.00 0 0.54 TYR 128 A 0.00 XXX 0 X 0.07 ASP 85 A
LYS 127 A 0.47 ASP 139 A 0.00 XXX 0 X 0.17 TYR 88 A
LYS 127 A 0.00 XXX 0 X 0.00 XXX 0 X 0.38 TYR 128 A
LYS 127 A 0.00 XXX 0 X 0.00 XXX 0 X 0.39 ASP 139 A
LYS 133 A 10.56 0 % -0.10 133 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.16 ASP 130 A
LYS 149 A 10.35 0 % -0.19 246 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.04 GLU 214 A
LYS 154 A 9.73 28 % -0.85 359 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.08 GLU 214 A
LYS 159 A 10.25 0 % -0.28 262 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.04 ASP 180 A
LYS 168 A 10.78 10 % -0.54 309 0.00 0 0.42 GLU 238 A 0.00 XXX 0 X 0.06 ASP 165 A
LYS 168 A 0.00 XXX 0 X 0.00 XXX 0 X 0.34 GLU 238 A
LYS 170 A 9.79 29 % -0.76 362 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.05 GLU 236 A
LYS 170 A 0.00 XXX 0 X 0.00 XXX 0 X 0.01 GLU 239 A
LYS 172 A 11.30 0 % -0.27 210 0.00 0 0.68 GLU 234 A 0.00 XXX 0 X 0.38 GLU 234 A
LYS 213 A 10.31 1 % -0.49 285 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.27 ASP 190 A
LYS 213 A 0.00 XXX 0 X 0.00 XXX 0 X 0.03 GLU 214 A
LYS 225 A 10.58 0 % -0.27 254 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.21 ASP 162 A
LYS 225 A 0.00 XXX 0 X 0.00 XXX 0 X 0.11 ASP 165 A
LYS 225 A 0.00 XXX 0 X 0.00 XXX 0 X 0.03 GLU 221 A
LYS 228 A 10.25 0 % -0.15 222 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.05 ASP 165 A
LYS 228 A 0.00 XXX 0 X 0.00 XXX 0 X -0.06 LYS 168 A
LYS 228 A 0.00 XXX 0 X 0.00 XXX 0 X -0.09 LYS 225 A
LYS 252 A 10.35 0 % -0.38 259 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.26 GLU 26 A
LYS 252 A 0.00 XXX 0 X 0.00 XXX 0 X 0.06 GLU 205 A
LYS 252 A 0.00 XXX 0 X 0.00 XXX 0 X -0.01 ARG 246 A
LYS 252 A 0.00 XXX 0 X 0.00 XXX 0 X -0.08 ARG 254 A
LYS 257 A 10.63 0 % -0.38 251 0.00 0 0.17 ASP 41 A 0.00 XXX 0 X 0.34 ASP 41 A
ARG 27 A 11.44 53 % -1.52 429 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.00 TYR 88 A
ARG 27 A 0.00 XXX 0 X 0.00 XXX 0 X 0.04 ASP 139 A
ARG 27 A 0.00 XXX 0 X 0.00 XXX 0 X 0.04 TYR 194 A
ARG 27 A 0.00 XXX 0 X 0.00 XXX 0 X 0.19 GLU 205 A
ARG 27 A 0.00 XXX 0 X 0.00 XXX 0 X 0.19 CYS 206 A
ARG 58 A 13.27 0 % -0.35 276 0.00 0 0.66 GLU 69 A 0.00 XXX 0 X 0.02 ASP 175 A
ARG 58 A 0.00 XXX 0 X 0.00 XXX 0 X 0.44 GLU 69 A
ARG 89 A 13.02 14 % -0.85 320 0.00 0 0.77 ASP 75 A 0.00 XXX 0 X 0.04 ASP 71 A
ARG 89 A 0.00 XXX 0 X 0.00 XXX 0 X 0.14 ASP 72 A
ARG 89 A 0.00 XXX 0 X 0.00 XXX 0 X 0.43 ASP 75 A
ARG 182 A 14.00 0 % -0.32 234 0.00 0 1.29 ASP 180 A 0.00 XXX 0 X 0.15 ASP 52 A
ARG 182 A 0.00 XXX 0 X 0.00 XXX 0 X 0.38 ASP 180 A
ARG 227 A 12.59 31 % -1.18 367 0.00 0 0.84 ASP 101 A 0.00 XXX 0 X 0.07 ASP 243 A
ARG 227 A 0.00 XXX 0 X 0.00 XXX 0 X 0.37 ASP 101 A
ARG 246 A 10.44 74 % -2.50 488 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.09 TYR 7 A
ARG 246 A 0.00 XXX 0 X 0.00 XXX 0 X 0.29 GLU 106 A
ARG 246 A 0.00 XXX 0 X 0.00 XXX 0 X 0.04 TYR 194 A
ARG 246 A 0.00 XXX 0 X 0.00 XXX 0 X 0.02 GLU 205 A
ARG 254 A 11.16 48 % -1.89 416 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.04 GLU 26 A
ARG 254 A 0.00 XXX 0 X 0.00 XXX 0 X 0.52 TYR 194 A
ARG 254 A 0.00 XXX 0 X 0.00 XXX 0 X 0.11 GLU 205 A
ARG 254 A 0.00 XXX 0 X 0.00 XXX 0 X -0.12 ARG 27 A
N+ 4 A 7.86 0 % -0.18 244 0.00 0 0.00 XXX 0 X 0.00 XXX 0 X 0.07 ASP 19 A
N+ 4 A 0.00 XXX 0 X 0.00 XXX 0 X -0.03 LYS 170 A
--------------------------------------------------------------------------------------------------------
SUMMARY OF THIS PREDICTION
Group pKa model-pKa ligand atom-type
ASP 19 A 3.87 3.80
ASP 32 A 3.58 3.80
ASP 34 A 3.33 3.80
ASP 41 A 2.04 3.80
ASP 52 A 3.50 3.80
ASP 71 A 1.68 3.80
ASP 72 A 4.21 3.80
ASP 75 A 3.33 3.80
ASP 85 A 3.82 3.80
ASP 101 A 3.14 3.80
ASP 110 A 2.44 3.80
ASP 130 A 3.59 3.80
ASP 139 A 2.36 3.80
ASP 162 A 3.45 3.80
ASP 165 A 3.84 3.80
ASP 175 A 3.92 3.80
ASP 180 A 2.35 3.80
ASP 190 A 3.98 3.80
ASP 243 A 2.52 3.80
GLU 14 A 3.84 4.50
GLU 26 A 4.50 4.50
GLU 69 A 4.88 4.50
GLU 106 A 5.12 4.50
GLU 117 A 3.18 4.50
GLU 187 A 4.83 4.50
GLU 205 A 3.96 4.50
GLU 214 A 3.75 4.50
GLU 221 A 3.89 4.50
GLU 234 A 3.64 4.50
GLU 236 A 3.87 4.50
GLU 238 A 3.22 4.50
GLU 239 A 5.15 4.50
HIS 10 A 6.39 6.50
HIS 15 A 6.26 6.50
HIS 17 A 5.54 6.50
HIS 36 A 6.74 6.50
HIS 64 A 3.47 6.50
HIS 94 A -2.19 6.50
HIS 96 A -0.62 6.50
HIS 107 A 6.50 6.50
HIS 119 A -4.89 6.50
HIS 122 A 3.58 6.50
CYS 206 A 11.58 9.00
TYR 7 A 13.35 10.00
TYR 40 A 10.36 10.00
TYR 51 A 13.81 10.00
TYR 88 A 12.05 10.00
TYR 114 A 11.04 10.00
TYR 128 A 10.78 10.00
TYR 191 A 12.05 10.00
TYR 194 A 13.92 10.00
LYS 9 A 10.55 10.50
LYS 18 A 10.26 10.50
LYS 24 A 10.24 10.50
LYS 39 A 10.26 10.50
LYS 45 A 10.51 10.50
LYS 76 A 11.27 10.50
LYS 80 A 10.34 10.50
LYS 111 A 10.74 10.50
LYS 112 A 9.97 10.50
LYS 113 A 10.22 10.50
LYS 127 A 12.13 10.50
LYS 133 A 10.56 10.50
LYS 149 A 10.35 10.50
LYS 154 A 9.73 10.50
LYS 159 A 10.25 10.50
LYS 168 A 10.78 10.50
LYS 170 A 9.79 10.50
LYS 172 A 11.30 10.50
LYS 213 A 10.31 10.50
LYS 225 A 10.58 10.50
LYS 228 A 10.25 10.50
LYS 252 A 10.35 10.50
LYS 257 A 10.63 10.50
ARG 27 A 11.44 12.50
ARG 58 A 13.27 12.50
ARG 89 A 13.02 12.50
ARG 182 A 14.00 12.50
ARG 227 A 12.59 12.50
ARG 246 A 10.44 12.50
ARG 254 A 11.16 12.50
N+ 4 A 7.86 8.00
--------------------------------------------------------------------------------------------------------
--------------------------------------------------------------------------------------------------------
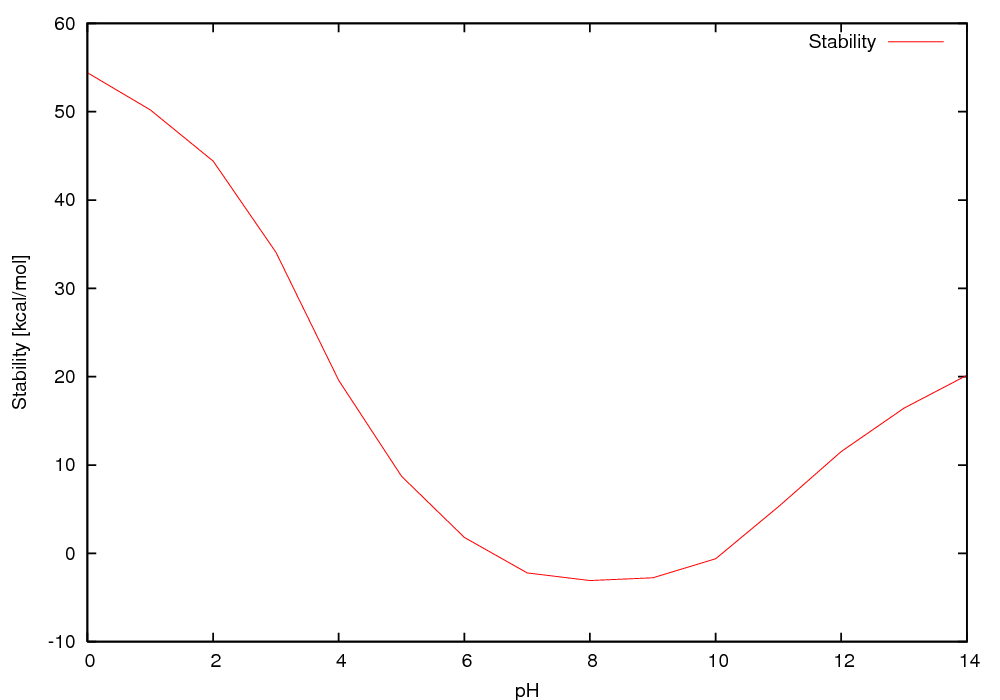
Free energy of folding (kcal/mol) as a function of pH (using neutral reference)
0.00 54.40
1.00 50.20
2.00 44.41
3.00 34.08
4.00 19.57
5.00 8.72
6.00 1.79
7.00 -2.22
8.00 -3.08
9.00 -2.77
10.00 -0.62
11.00 5.28
12.00 11.53
13.00 16.44
14.00 20.16
The pH of optimum stability is 8.2 for which the free energy is -3.1 kcal/mol at 298K
The free energy is within 80 % of maximum at pH 7.2 to 9.2
The free energy is negative in the range 6.4 - 10.1
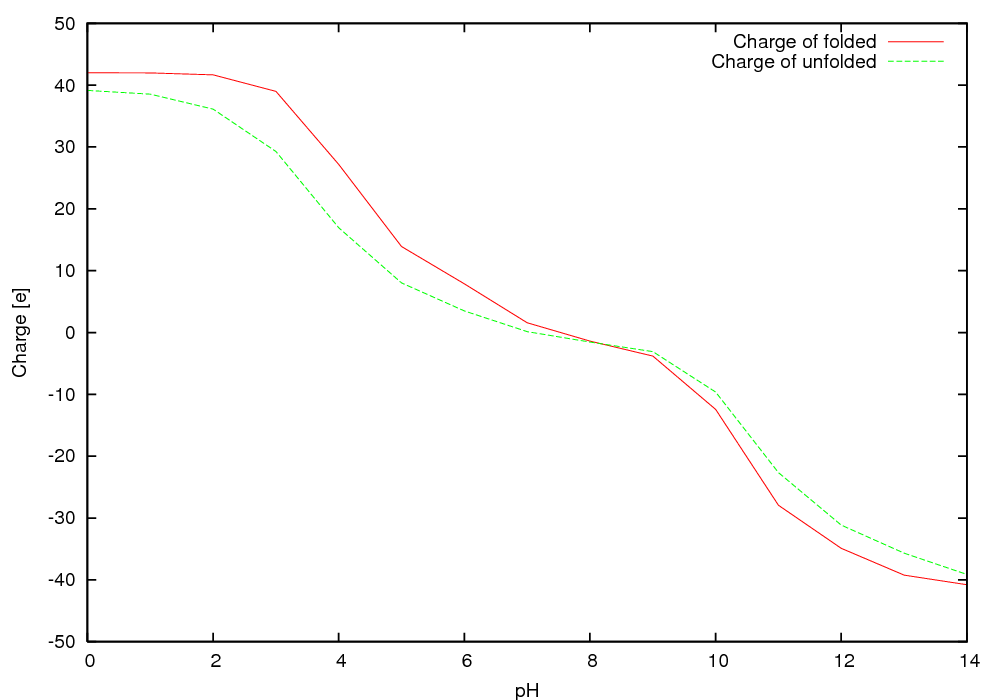
Protein charge of folded and unfolded state as a function of pH
pH unfolded folded
0.00 42.00 39.15
1.00 41.97 38.55
2.00 41.66 36.12
3.00 39.00 29.26
4.00 27.19 16.93
5.00 13.91 8.01
6.00 7.86 3.49
7.00 1.58 0.14
8.00 -1.40 -1.54
9.00 -3.81 -3.10
10.00 -12.44 -9.63
11.00 -27.95 -22.67
12.00 -34.90 -31.15
13.00 -39.24 -35.69
14.00 -40.78 -39.12
The pI is 7.10 (folded) and 7.40 (unfolded)
 EPS format |
 EPS format |